Developmental Genetics
Control of spatial laser beam formation in the development of multi-view light sheet microscopy
The Developmental Genetics (DG) group studies the regulation of dynamic cell behaviors in the development of the Drosophila melanogaster embryo [1]. The tools used in these studies include molecular cell biology, genetics, and high-resolution imaging. The Drosophila embryo has a rather large size (500 µm long, 200 µm across), which provides a challenge for conventional microscopic techniques. The overall aim of our project is to design and build a microscope with high spatio-temporal resolution based on a selective plane illumination microscopy platform (SPIM or light sheet microscopy). SPIM is based on optical sectioning by illuminating the sample with a thin light sheet orthogonal to the detection axis (Fig. 1a). The sectioning capability in SPIM is defined by the thickness of the light sheet. A key area of the SPIM research is to produce an optimal light sheet which covers a large field of view with a high spatial resolution. Different methods, including specific spatial beam structures (e.g. Bessel-, Airy-, Gaussian beams) or tiling methods, have been advised to address this problem, but these are often difficult to apply for biologists with little training in optics or physics. To produce optimized light sheets for specific biological applications we have developed a software called Structured SPIM (SSPIM), which provides an open-source, user-friendly and compact tool- box for beam shaping that can generate digital patterns for a wide range of structures for shaping optical beams [2]. SSPIM can produce Gaussian, Bessel and Airy beams (Fig. 1b-g) by simple control of a Spatial Light Modulator (SLM). SSPIM is also able to instruct patterns for incoherent and coherent array beam formation and beam tiling (Fig. 2) [2]. SSPIM provides a key component of our current development of a dual-view tiling OpenSPIM system with which we are now able to record the whole functional 3D image of dynamic cell behaviors in Drosophila gastrulation at subcellular resolution.
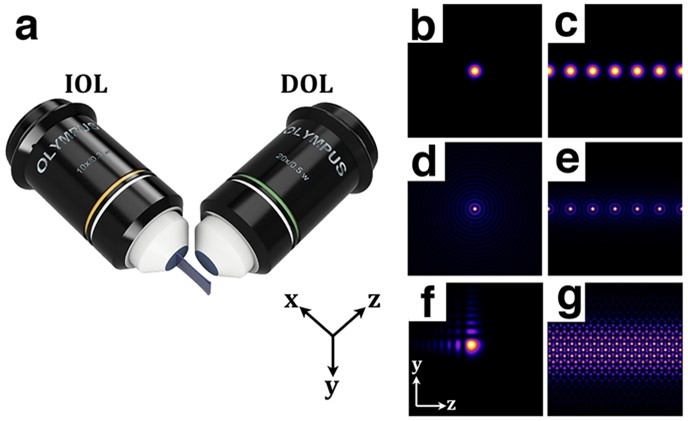
Figure 1: Schematic of the SPIM and example of illumination beams. (a) Principle of SPIM: the illumination objective lens (IOL) excites a thin sheet (x-y plane); the emitted light is collected with the detection objective lens (DOL) in the orthogonal direction (z axis). (b, d) Transverse intensity profiles of single Gaussian and Bessel with radial symmetry and (f) 2D Airy beams with asymmetrical intensity profile. (c,e,g) For better resolution and contrast, Gaussian (c) and Bessel array beams (e) and Lattice beams (g) can be applied into the SPIM.
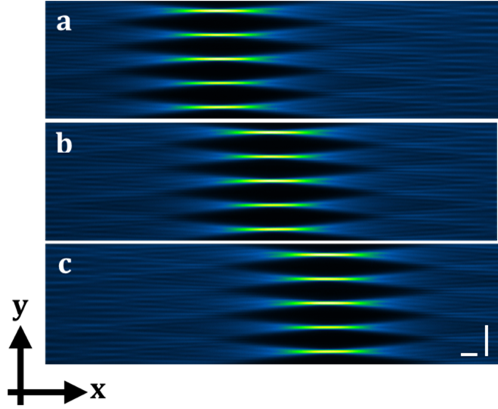
Figure 2: Tiling method. (a-c) show the measured intensity profile of the tiled Gaussian array beam through the dye solution. The intensity of the green color indicates the thinnest light sheets and thus the best spatial resolution. The tiling method allows the sequential illumination of the sample with the best spatial resolution of the light sheets. Tiling is achieved by applying SSPIM to control a spatial light modulator (SLM). Scale bars: 40 μm.
Airyscan super-resolution confocal microscopy reveals dynamics of cell division proteins
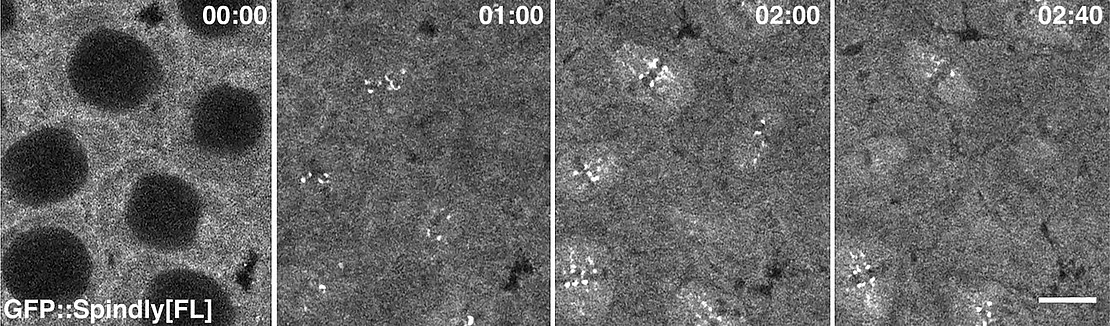
A second research topic in the DG group is the analysis of adaptor proteins of the motor protein Dynein and their role in controlling the specificity and activity of Dynein-mediated transport along microtubules. In a recent study, we have investigated the role of the Dynein adaptor Spindly, which was originally identified as a specific regulator of Dynein activity during cell division. While some details of Spindly function in cell divisions have been worked out in other systems, the function of Spindly within the context of an organism had not yet been addressed. Knock-down of Spindly protein by RNA Interference (RNAi) demonstrated that Spindly is required for mitotic cell divisions in the embryo and for cell migration of follicle cells in the ovary [3]. This genetic background was used to study the localization of Spindly using transgenic proteins tagged with green fluorescent protein (GFP). We showed for the first time the dynamic distribution of Spindly protein in a living organism (Fig. 3). The Spindly-GFP system allowed the analysis of Spindly protein domains for its localization and function revealing that the carboxy-terminal region (DTR) controls Spindly localization in a cell-type specific manner. While the CTR is dispensable for Spindly localization and function in the embryo, in larval founder cells of nervous system the CTR is necessary for Spindly localization. We also discovered, that increasing the level of Spindly in the female germ line results in lethality and alters the morphology of the egg. To determine whether Spindly plays a role in post-mitotic cells, we altered Spindly protein levels in migrating cells and found that ovarian border cell migration is sensitive to the levels of Spindly protein. In summary our study provides the first demonstration of Spindly function and dynamic distribution in a living organism and uncovers novel functions of the protein revealing a differential, functional requirement for its carboxy-terminal region in Drosophila.
References
[1] Winklbauer R and Muller HA (2011). Mesoderm layer formation in Xenopus and Drosophila gastrulation. Phys Biol 8(4):045001.
[2] Aakhte, M, Akhlaghi, EA and HAJ Müller (2018). SSPIM: a beam shaping toolbox for structured selective plane illumination microscopy. Sci. Reports 8, 10067.
[3] Clemente GD, Hannaford MR, Beati H, KappK, Januschke J, Griffis ER and Müller HAJ (2018). Requirements of the Dynein-adaptor Spindly for Mitotic and Post-Mitotic Functions in Drosophila. J Dev Biol. 6 (2), 9.
Contact
Prof. Dr. Arno Müller
full member

- Telephone
- +49 561 804-4725
- h.a.muller[at]uni-kassel[dot]de
- Location
- Universität Kassel
Fachbereich 10 - Naturwissenschaften & Mathematik
Institut für Biologie
Heinrich-Plett-Straße 40
34132 Kassel
- Room
- IBC, 2403
