Microbiology
The Division of Microbiology led by Professor Schaffrath uses yeast model systems to study protein and RNA modifications that influence mRNA translation, protein synthesis and cell growth. One of the protein modifications of interest is called urmylation. Here, the ubiquitin-like modifier Urm1 is transferred to a target protein (e.g., Ahp1) via a lysine-directed conjugation reaction that is conserved from yeast to human cells. The function and fate of urmylated proteins are still unclear, which is why the group further investigates this non-canonical Urm1 conjugation pathway.
Thiol peroxidases and peroxiredoxins ensure cellular redox homeostasis and are thus crucial for the protection of cells against reactive oxygen species (ROS) and oxidative stress. Their importance is evidenced by disease formation and premature aging in cells (including our own) associated with defects in thiol peroxidases. Ahp1 (alkyl hydroperoxidase 1) is a 2-Cys peroxiredoxin from yeast that maintains two redox-active thiols (C31, C62) (Fig. 1) critical for ROS detoxification. During catalysis, Ahp1 forms a homo-dimer and reduces ROS with its peroxidatic Cys (CP: C62) residues that get oxidized to a sulfenic acid (-SOH). Next, the sulfenylated CPs are disulfide-linked to the resolving Cys (CR: C31) residues from opposite Ahp1 subunits in the dimer (Fig. 1). Upon reduction by thioredoxin, the homo-dimer becomes repaired for another Ahp1 peroxidatic cycle (Fig. 1).
Interestingly, redox stress triggers post-translational modification of Ahp1 including urmylation, a conjugation to the ubiquitin-like protein Urm1 (Fig. 1). Urm1 not only modifies proteins but also acts as a sulfur-donor for tRNA thiolation together with Elongator, a tRNA anticodon modifier complex. Importantly, tRNA thiolation and ROS dependent Ahp1 urmylation both require Urm1 to be thiocarboxylated at its C-terminus (Urm1-COSH). This suggests that the two Urm1 functions are linked by S-transfer coupling both to oxidative stress. Consistently, several groups including our own have shown that redox stressors and thiol-reactive agents (H2O2, diamide) trigger protein urmylation. Indeed, among Urm1 targets identified from yeast, fungi, flies and human cells are factors of the oxidative stress response.
Nonetheless, a specific function for urmylation of Ahp1 remains elusive. Apparently, Urm1 is not a (ubiquitin-like) tag for degradation but may otherwise affect the peroxiredoxin. In line with this, it was shown that Ahp1 is urmylated at Lys residue K32, which is next to the redox-active CR (C31) (Fig. 1). Hence, Urm1 conjugation may interfere with Ahp1 through conformational changes near the redox-active thiol center (Fig. 1) that is critical for catalysis.
Having established under DFG (SCHA750/15-1) support tools and protocols for the diagnosis of urmylation, the Schaffrath group currently addresses mechanistic aspects of urmylation and the relevance of Urm1 conjugation (SCHA750/15-2) for the cell in cooperation with the Max Planck Research group led by Dr Glatt (Jagiellonian University Krakow, Poland). How exactly sulfur (Urm1-COSH) is used during the urmylation reaction is unclear, making it crucial to study its fate post-urmylation. Also, whether and how urmylation of Ahp1 may affect the thiol-based redox-active switch, which is important for the peroxiredoxin to function in the response to ROS, are key questions in need for answers. In particular, the following ill-defined aspects of the urmylation pathway are in the focus of the group:
- the role of the activated sulfur in Urm1-COSH for the Urm1 conjugation pathway
- the fate the activated S-species derived from Urm1-COSH faces post-urmylation
- the role of urmylation for Ahp1 performance in the ROS response
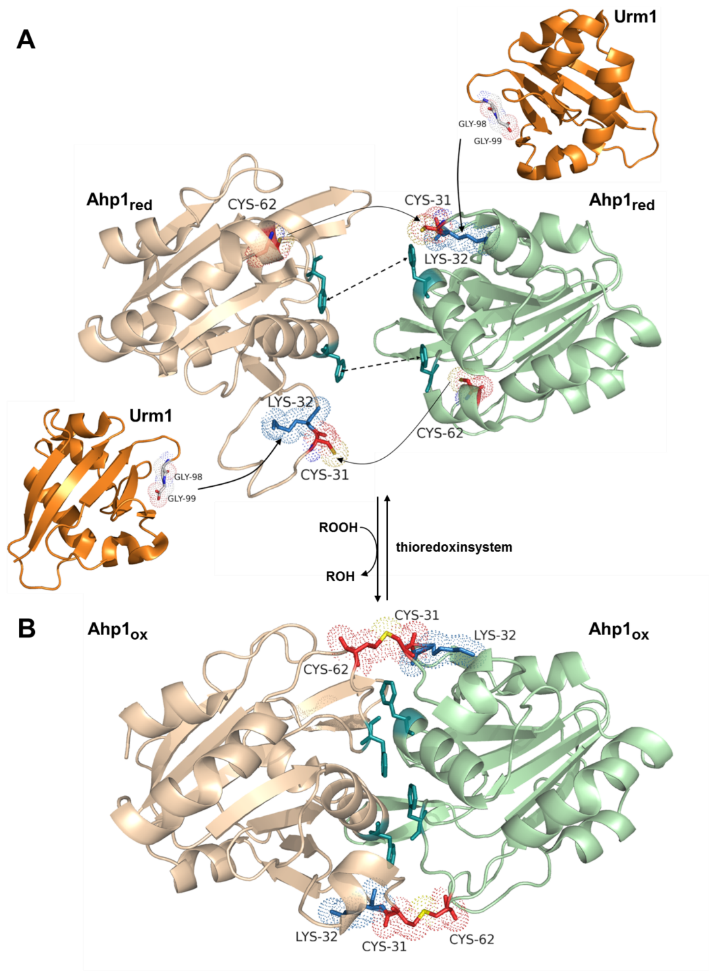
Fig. 1. Structure and urmylation of the
peroxiredoxin Ahp1.
(A) Overview of reduced Ahp1 (Ahp1red) subunits (beige/green) (PDB: 4DSR) with details on the urmylation site (Lys-32, blue) and the cysteine (red) based thiols (yellow) in CR (Cys-31) and CP (Cys-62). Urmylation uses a highly conserved di-glycine motif of Urm1 (orange) (PDB: 2QJL) to form an isopeptide bond with Lys-32 of the respective Ahp1 subunit. The interface Phe residues (dark green), which provide the hydrophobic interactions (dotted line) between the two subunits, are highlighted. (B) After oxidation of Ahp1 (Ahp1ox), the dimer (PDB: 4DSQ) undergoes a conformational change and a disulfide (yellow) is formed between the CP of one (beige) and the CR of another (green) subunit. It has been shown that the hydrophobic Phe side chains bring the two subunits into the correct orientation to each other, regardless of redox status. Crystal structures: J Biol Chem 287: 17077ff. Illustration created with The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC.
References
Jüdes A, Bruch A, Klassen R, Helm M, Schaffrath R (2016) Sulfur transfer and activation by ubiquitin-like modifier system Uba4•Urm1 link protein urmylation and tRNA thiolation in yeast. Microb Cell 3, 554-564. doi 10.15698/mic2016.11.539
Jüdes A, Ebert F, Bär C, Thüring KL, Harrer A, Klassen R, Helm M, Stark MJR, Schaffrath R (2015) Urmylation and tRNA thiolation functions of ubiquitin-like Uba4•Urm1 systems are conserved from yeast to man. FEBS Lett 589, 904-909. doi 10.1016/j.febslet.2015.02.024
Prof. Dr. Raffael Schaffrath
full member

- Telephone
- +49 561 804-4175
- schaffrath[at]uni-kassel[dot]de
- Location
- Universität Kassel
Fachbereich 10 - Naturwissenschaften & Mathematik
Institut für Biologie
Heinrich-Plett-Straße 40
34132 Kassel
- Room
- 2104
