Project 10
P10: Multiscale-multicomponent theoretical description of biological oscillators and clocks: from subcellular-level simulations to network modeling
Collaborators
- PI Dr. Kapp & PI Prof. Dr. Müller (Biology) (öffnet neues Fenster)
- PI Prof. Dr. Mayer (Biology) (öffnet neues Fenster)
- PI Prof. Dr. Seiler (Mathematics) (öffnet neues Fenster)
- PI Prof. Dr. Stengl (Biology) (öffnet neues Fenster)
- PI Prof. Dr. Stursberg (Electrical engineering/Informatics) (öffnet neues Fenster)
Project description
Long-term electrophysiological signals measured from cockroach brains in vivo revealed biological rhythms covering time scales from milliseconds to days [1].
The identification and understanding of biological clocks/oscillators with different periods and their crosstalk in organisms requires to develop new modeling tools and to also apply methods from other scientific disciplines. Our interdisciplinary theoretical group has experience in the analysis of electrophysiological signals [1,2], in the development of models based on experimental or simulation data [2-4], in Langevin- [2] and Monte-Carlo [5] simulations as well as in ab-initio and large-scale atomistic simulations, both in condensed matter [3,4] and in biological systems [6,7].
Objectives of our Project: we will develop and apply theoretical methods to answer the following questions: (1) How can the topology (neural structure) of a multicellular circadian clock network regulating processes on the time scale of hours that are triggered by transient neuropeptide release (timescales: milliseconds to seconds) be inferred from multiscale electrophysiological recordings? (2) How do single circadian-clock neurons, being mainly driven by stochastic biomolecular processes, generate and orchestrate synchronized multiscale oscillations? (3) How do rhythmically occurring membrane invaginations during embryogenesis couple to biochemical oscillations of cell cycle clocks? (3) How robust and how adaptive are biological clocks/oscillators? Are simple terrestrial organisms able to adapt themselves to non-24 h day-night-cycles of other planets of our solar system or even exoplanets?
In the framework of the RTG we will closely collaborate with most other involved groups.
[1] Jarukanont, D., et al., Vesicle Motion during Sustained Exocytosis in Chromaffin Cells: Numerical Model Based on Amperometric Measurements, PLoS ONE 10, e0144045 (2015). https://doi.org/10.1371/journal.pone.0144045
[2] Rojas, P., et al., Beyond spikes: Multiscale computational analysis of in vivo long-term recordings in the cockroach circadian clock. Network Neuroscience 3, 944-968 (2019).https://doi.org/10.1162/netn_a_00106
[3] Zijlstra, E. S., et al., Fractional Diffusion in Silicon. Advanced Materials, 25, 5605–5608 (2013).https://doi.org/10.1002/adma201302559
[4] Bauerhenne, B., et al., Self-Learning Method for Construction of Analytical Interatomic Potentials to Describe Laser-Excited Materials. Physical Review Letters, 124, 085501(2020).https://doi.org/10.1103/PhysRevLett.124.085501
[5] Ojeda-May, P., & Garcia, M. E., Electric Field-Driven Disruption of a Native β-Sheet Protein Conformation and Generation of a Helix-Structure. Biophysical Journal, 99, 595–599 (2010). https://doi.org/10.1016/j.bpj.2010.04.040
[6] Reuter, B., et al., Generalized Markov State Modeling Method for Nonequilibrium Biomolecular Dynamics: Exemplified on Amyloid β Conformational Dynamics Driven by an Oscillating Electric Field, J. Chem. Theory Comput., 14, 3579-3594 (2018). https://doi.org/10.1021/acs.jctc.8b00079
[7] Arbeitman, C. R., et al., The SARS-CoV-2 spike protein is vulnerable to moderate electric fields. Nature Communications12, 5407 (2021).https://doi.org/10.1038/s41467-021-25478-7
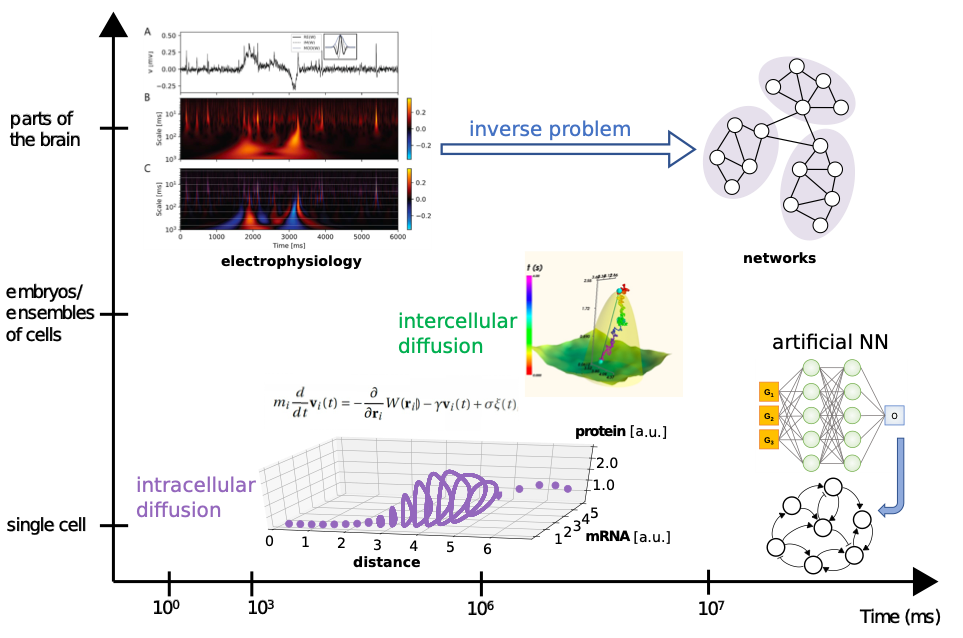
Figure 1: summary of the time scales, systems and methods considered in the present project. We plan to perform different types of simulations and use Machine-Learning based methods for data analysis and for theoretical predictions on a variety of organisms.
